Dr Julie Aspden
- Position
- Associate Professor of RNA biology
- Areas of expertise
- RNA, mRNA translation, long non-coding RNAs, ribosomes, RNA-binding proteins, Drosophila melanogaster, spermatogenesis, neuronal differentiation
- j.aspden@leeds.ac.uk
- Phone
- 0113 343 5646
- Location
- Garstang 9.22
- Faculty
- Biological Sciences
Introduction
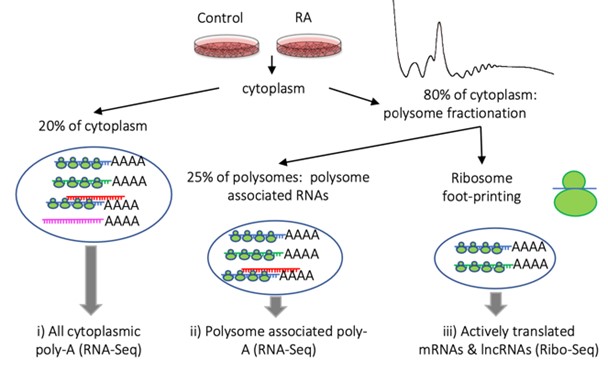
My group are interested in RNA and its role in gene expression. We broadly address questions on the regulation of mRNA translation, non-coding RNA function and the role of specific RNA-protein complexes. Using a combination of biochemistry, genomics, molecular biology and genetics to study RNAs both in Drosophila melanogaster and several mammalian systems. To understand what is being translated we use ribosome profiling (Ribo-Seq) to sequence the fragments of RNA bound by translating ribosomes. I previously developed an adaptation of this approach to identify novel regions of translation, Poly-Ribo-Seq.
Many of the regulatory processes and RNA-binding proteins are highly conserved between Drosophila and mammals. Our research focus is of general interest and importance because disruptions to RNA-protein interactions and translational regulation play significant roles in a variety of cancers and other disorders (e.g. spinal muscular atrophy) and many non-coding RNAs show correlations with neuronal disease phenotypes but their role in cellular mis-function has not been elucidated.
Current major projects
- Specialised ribosomes
- Discovery of novel open reading frames and production of micropeptides
- Function of cytoplasmic lncRNAs during neuronal differentiation
- mRNA translation regulation during spermatogenesis
Detailed research programme
Specialised ribosomes
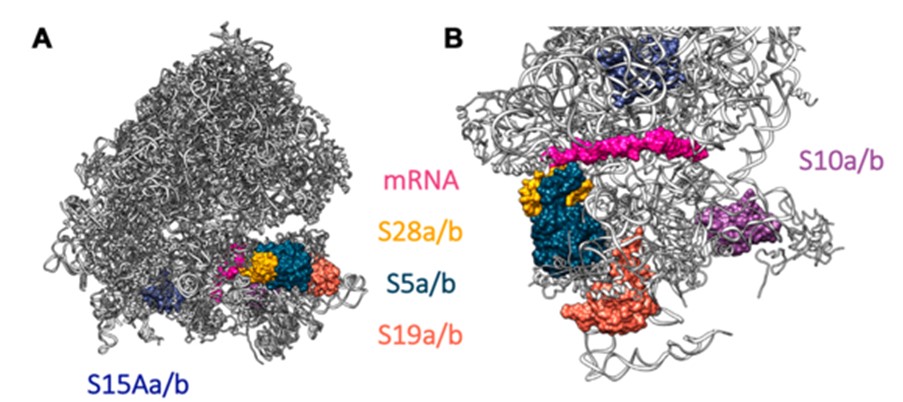
Changes in protein composition generated through ribosome heterogeneity facilitate the ability of ribosomes to selectively translate specific mRNAs and ORFs. However, the underlying mechanisms by which these 'specialised ribosomes' regulate translation are unknown. In collaboration my group are seeking to understand the evolution, function and structure of specialised ribosomes. This will impact our understanding of pathologies caused by deregulation of translation, including ribosomopathies and cancer, and future biotechnology applications.
We have previously identified ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching events using quantitative mass spectrometry and cryo-EM of ribosomes (in collaboration with Dr Juan Fontana) from different Drosophila tissues (Hopes et al NAR 2022).
Discovery of novel open reading frames and production of micro-peptides
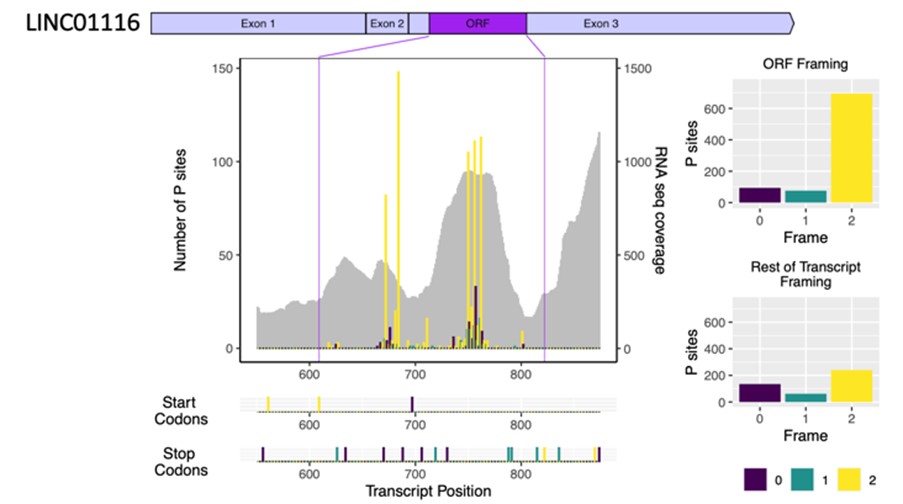
The discovery that long non-coding RNAs (lncRNAs) can be translated to produce non-canonical micro-peptides has revealed an unexplored extension to the human proteome. Those novel micro-peptides that have been characterised have important roles in cellular processes, particularly in membranes (e.g. calcium transport) and mitochondria (e.g. oxidative phosphorylation).
This recently identified source of novel peptides is particularly relevant in neuronal cells, where lncRNAs are highly enriched. My group has previously identified 45 novel micropeptides translated from lncRNAs during neuronal differentiation. These translated lncRNAs contribute to neuronal differentiation but the function of the resulting micro-peptides has yet to be determined. My group is interested in understanding the role of these translated lncRNAs during neuronal differentiation of cortical organoids (in collaboration with Dr James Poulter). This work provides insight into the causes of neuronal diseases and reveal how disease associated lncRNAs are contributing to neuronal pathology.
Work is also underway to understand the evolution of these novel ORFs and how they represent an important step on the evolution of novel protein coding genes (in collaboration with Dr Mary O’Connell).
mRNA translation regulation during spermatogenesis
My group is interested in understanding how mRNA translation is regulated during spermatogenesis when gene expression is tightly regulated post-transcriptionally. Recently we have characterised the signal transduction and activation of RNA (STAR) protein RNA-binding protein ‘Held-out-wings’, which is an ortholog of human quaking. Using RIP-Seq we have identified which identified 121 novel transcripts bound by HOW(S) in germ stem cells and spermatogonia, many with signal transduction functions. (A/G/U)CUAAC motifs were enriched in 3'-UTRs and GCG(A/U)G in 5'-UTRs. Using fluorescence anisotropy (in collaboration with Thomas Edwards) we have shown that HOW binds with high-affinity to sites containing CUAAC motifs from lola and hipk mRNAs. This work provides new insight into STAR protein-RNA interactions and functions in spermatogenesis (in collaboration with Dr Amanda Bretman).