Professor Alan Berry
- Position
- Professor of Molecular Enzymology
- Areas of expertise
- Enzymology; Protein engineering; Directed Evolution; Biocatalysis; Enzyme mechanisms
- a.berry@leeds.ac.uk
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
Introduction
We have a wide range of interests in synthetic biology and its applications in generating novel proteins and enzymes using rational design and directed evolution. Our work spans biochemistry and chemistry with an emphasis on the enzymes that make and break carbon-carbon bonds, particularly aldolases, terpene synthases and polyketide synthases. We have used the modern methods of protein engineering and directed evolution to alter the specificity of enzyme reactions, the stability of enzymes and their stereochemistry, and structural techniques such as X-ray crystallography, cryo-electron microscopy and NMR spectroscopy with enzyme kinetics and advanced reaction modelling to characterise the resulting enzymes.
Current major projects
- Enzymology of polyketide and polyether synthases
- Investigating and engineering the specificity of terpene synthases
- Creating new enzymes containing non-natural amino acids
Detailed research programme
Characterising and engineering polyketide synthases / non-ribosomal peptide synthases.
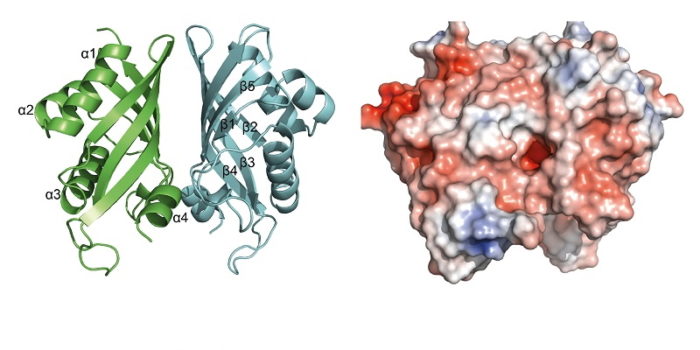
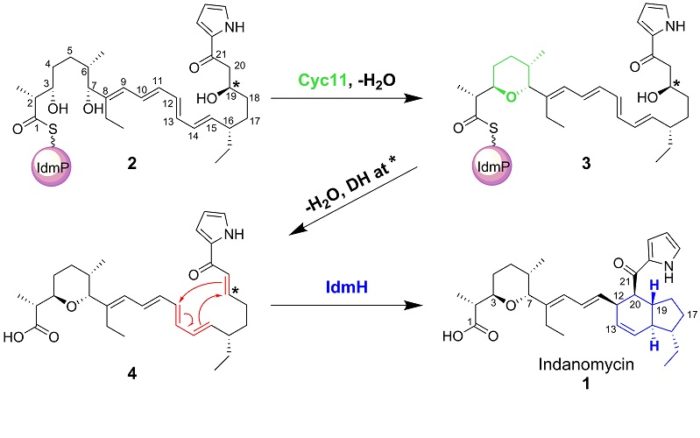
The modular PKS/NRPSs offer great potential to engineer the specificity of product production by altering the individual modules in the enzyme. We are investigating the structure and mechanism of the PKS/NRPS systems responsible for the biosynthesis of a number of compounds. Polyketides also often undergo further ‘tailoring’ reactions and we are also studying a number of these. For example, we have recently solved the structure and modelled the reaction of the Diels-Alderase involved in the biosynthesis of indanomycin.
Investigating and engineering the specificity of terpene synthases
A number of complex secondary metabolites are generated from isoprene units and these molecules have important uses as pharmaceuticals, flavours and pigments. These molecules are generated from two simple precursors, dimethyl allyl diphosphate, and isopentenyl diphosphate and are built into the complex structures by synthases which join these precursors together and then shape and cyclise the intermediates into the final structures. Our work on selinadiene synthase aims to determine which residues in the enzyme active site are responsible for various steps in the reaction mechanism and to use that knowledge to re-engineer the enzyme to produce a wider range of terpene products.
Creating new enzymes containing non-natural amino acids
Aldolases catalyse the condensation of aldehydes with ketones and offer great potential in biotechnology as efficient, green catalysts. However their properties can also cause problems - for example limited substrate range and stability. We have demonstrated the alteration of stability and specificity of a number of aldolases. Excitingly we have also engineered/evolved aldolases to alter their stereochemistry - giving the possibility of tailoring the specificity and stereochemical outcome of the reaction. Other recent work has shown that unnatural amino acids can be inserted into the enzyme active site and that activity can be retained and that new activities that are dependent on the location of an unnatural amino acid in the aldolase active site can be generated.