Dr Anastasia Zhuravleva
- Position
- Lecturer
- Areas of expertise
- NMR; structural biology; protein folding; molecular chaperones; stress sensors
- Location
- 6.01 Miall
- Faculty
- Biological Sciences
- School
- Mollecular and Cellular Biology
- Website
- ORCID
Introduction
Study and manipulation of protein homeostasis and the stress response raises the possibility of altering many pathological processes, including viral and bacterial infections, neurodegenerative diseases, diabetes and cancer. Complex multicomponent networks of stress sensors, molecular chaperones and degradation enzymes enable real-time adaptive adjustments of the cellular folding capacity under physiological and pathological stresses. My research focuses on the mechanistic understanding of the multicomponent protein quality control (PQC) system, including investigation of the complex relationship between structure, conformational flexibility, and function of individual PQC components, their interactions with each other and unfolded protein substrates as well as post-translation fine-tuning of the PQC networks in health and disease.
Current major projects
- Molecular mechanisms of activation for the endoplasmic reticulum stress sensor Ire1 and chaperone BiP
- How do post-translational modifications control Hsp70 chaperone functions?
- Molecular mechanism of action for the chaperone SurA from Gram-negative bacteria
- Integrative structural biology approaches for characterization of challenging biological systems
Detailed research programme
How is the BiP chaperone cycle fine-tuned post-translationally?
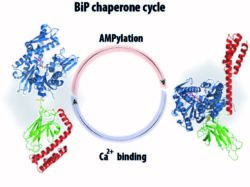 The understanding of how folding capacity in the ER is reversibly adjusted under physiological and pathological stresses, is of great biological and biomedical interest. Our research aims in characterization of molecular mechanisms for post-translational regulation for a key ER molecular chaperone BiP. Exploiting a methyl NMR approach, we have revealed key molecular features of the chaperone cycle of unmodified BiP and its alterations upon AMPylation (eLife 2017) and binding to Ca2+. Our results suggest that the thermodynamic distribution in the BiP conformational ensemble defines the level of BiP activity. In turn, covalent post-translational modifications (e.g., AMPylation) and binding to small metabolites and ions (e.g., Ca2+) perturb the BiP conformational ensemble and, thus, enable fine-tuning of BiP functions.
The understanding of how folding capacity in the ER is reversibly adjusted under physiological and pathological stresses, is of great biological and biomedical interest. Our research aims in characterization of molecular mechanisms for post-translational regulation for a key ER molecular chaperone BiP. Exploiting a methyl NMR approach, we have revealed key molecular features of the chaperone cycle of unmodified BiP and its alterations upon AMPylation (eLife 2017) and binding to Ca2+. Our results suggest that the thermodynamic distribution in the BiP conformational ensemble defines the level of BiP activity. In turn, covalent post-translational modifications (e.g., AMPylation) and binding to small metabolites and ions (e.g., Ca2+) perturb the BiP conformational ensemble and, thus, enable fine-tuning of BiP functions.
How does the molecular chaperone BiP control activation of the ER stress sensor Ire1?
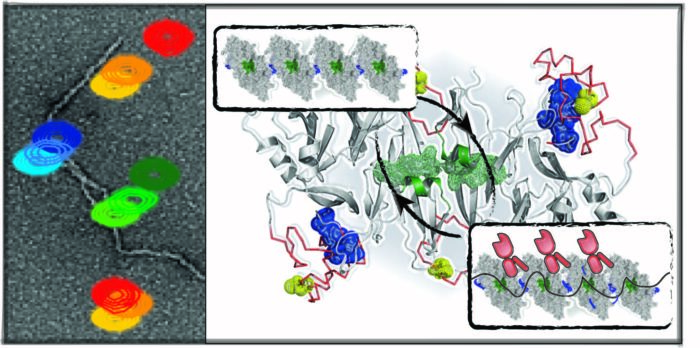
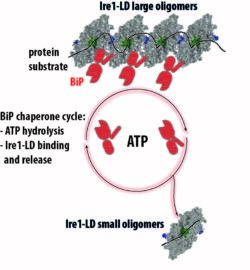 The kinase/endonuclease inositol-requiring enzyme 1 (Ire1), the evolutionarily oldest and conserved ER stress sensor, senses accumulation of unfolded proteins in the ER and transfers stress signals from the ER lumen. Using solution NMR and other biophysical techniques, we have demonstrated that in the absence of stress Ire1 luminal domain (LD) exists as low order oligomeric species, while interactions with unfolding protein substrates result in high-order oligomerization of Ire1-LD, triggering the stress response. In turn, BiP controls the Ire1 activation cascade through dynamic, ATP-dependent interactions between BiP and large Ire1-LD oligomers, resulting in their destabilization. Next, we aim to elucidate molecular mechanisms by which BiP and unfolded protein substrates orchestrate the Ire1 activation cascade in health and disease.
The kinase/endonuclease inositol-requiring enzyme 1 (Ire1), the evolutionarily oldest and conserved ER stress sensor, senses accumulation of unfolded proteins in the ER and transfers stress signals from the ER lumen. Using solution NMR and other biophysical techniques, we have demonstrated that in the absence of stress Ire1 luminal domain (LD) exists as low order oligomeric species, while interactions with unfolding protein substrates result in high-order oligomerization of Ire1-LD, triggering the stress response. In turn, BiP controls the Ire1 activation cascade through dynamic, ATP-dependent interactions between BiP and large Ire1-LD oligomers, resulting in their destabilization. Next, we aim to elucidate molecular mechanisms by which BiP and unfolded protein substrates orchestrate the Ire1 activation cascade in health and disease.
Molecular mechanisms of the bacterial chaperone SurA
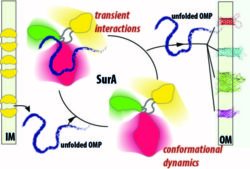 SurA is the conserved major chaperone of outer membrane proteins (OMPs) in the periplasm of Gram-negative bacteria, and plays a key role in cell envelope homeostasis and virulence. A remarkable feature of SurA (and other periplasmic chaperones) is that they assist the folding of OMPs and prevent their aggregation without consumption of ATP or assistance by co-chaperones. In collaboration with Sheena Radford (Astbury Centre for Structural Biology), we are exploiting the latest developments in solution NMR and other biophysical and computational techniques to characterize with atomic details the dynamic conformational landscape of SurA and elucidate the molecular basis of SurA binding and specificity to different OMPs.
SurA is the conserved major chaperone of outer membrane proteins (OMPs) in the periplasm of Gram-negative bacteria, and plays a key role in cell envelope homeostasis and virulence. A remarkable feature of SurA (and other periplasmic chaperones) is that they assist the folding of OMPs and prevent their aggregation without consumption of ATP or assistance by co-chaperones. In collaboration with Sheena Radford (Astbury Centre for Structural Biology), we are exploiting the latest developments in solution NMR and other biophysical and computational techniques to characterize with atomic details the dynamic conformational landscape of SurA and elucidate the molecular basis of SurA binding and specificity to different OMPs.
High-resolution hydrogen/deuterium exchange for challenging biomedical systems
Hydrogen/deuterium exchange (HDX) is a unique technique providing detailed information about structure and conformational flexibility of protein systems in solution and effects of drug binding, protein-protein interactions (PPIs), disease-related mutations and aggregations. To fully benefit from this approach, we have integrated DMSO-quenched HDX with the latest developments in fast multidimensional NMR to allow HDX rates to be obtained with single-residue resolution and using physiological