Professor Andrew Macdonald
- Position
- Professor of Tumour Virology
- Areas of expertise
- Cancer, Signal Transduction, Viruses, Therapeutics
- Location
- Garstang 8.53e
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
- Website
- ORCID
Introduction
The work undertaken in my group is truly interdisciplinary, traversing the disciplines of cell biology, structural biology and virology. We study DNA tumour viruses from the Papillomavirus and Polyomavirus families and are fascinated by the ways in which these viruses are able to infect and re-wire a host cell to enable virus replication, persistence and ultimately to cause pathogenesis. By studying the virus – host interactions that occur during infection we hope to leverage this information to identify new targets for therapeutic intervention.
Current major projects
- Expanding our knowledge of the tumour suppressors and oncogenes in virus-driven cancers
- How do RNA interaction networks regulate epithelial cancers?
- Understanding the molecular basis for virus entry
- Targeted therapeutics for virus infection
Detailed research programme
Expanding our knowledge of the tumour suppressors and oncogenes in virus-driven cancers
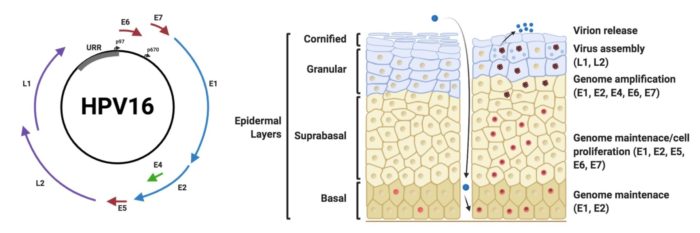
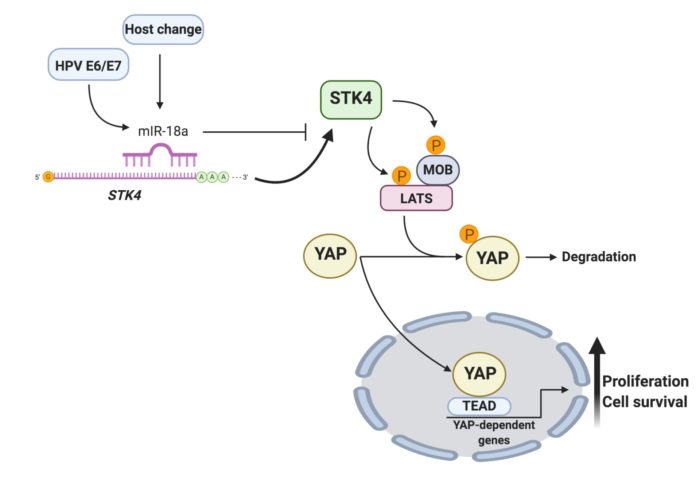
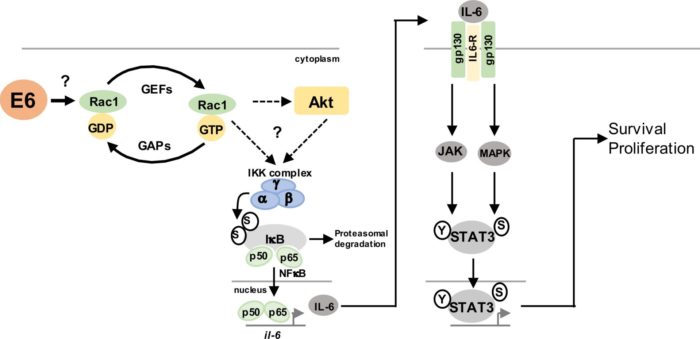
Oncogenic viruses are effective model systems to dissect the repertoire of host tumour suppressors and oncogenes in cancer. We have effectively used HPV and Merkel cell polyomavirus (MCV) to identify and characterise a number of established and newly emerging transformation mechanisms including the JAK-STAT3, Hippo and the p38 MAPK pathways. Currently we are working on a number of novel host factors previously not linked to transformation and are determining the molecular basis for their function in cancer.
How do RNA interaction networks regulate epithelial cancers?
Non-coding RNA molecules play an essential role in regulating cellular homeostasis. We have identified a number of oncogenic microRNAs that are essential for cervical cancer cell growth and survival and have mapped the cellular pathways that they regulate. Our ambition is to analyse the networks of these non-coding RNAs in epithelial cancers at the systems level, to truly appreciate their impact on cancer cell biology. Towards this, we work closely with clinical colleagues to amass tumour biopsy samples from which we can perform large scale RNA-Sequencing to better interrogate gene expression. Ultimately, we wish to identify novel mechanisms of cancer cell regulation.
Understanding the molecular basis for virus entry.
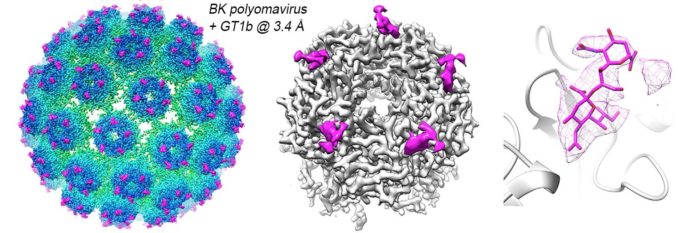
Virus capsids are multifunctional molecular machines that are essential to infection. Capsids must exist on a physico-chemical knife edge: they must be stable enough to protect the genome in harsh external environments, but fragile enough to release their genome at the right moment. They must interact with the target cell, select the appropriate entry route(s) and undergo structural transitions during entry and/or trafficking, to ultimately allow the genome to be captured by replication machineries, all whilst evading the host’s immune responses. We are using the human BK polyomavirus as a model system to understand non-enveloped virus entry mechanisms. We use a powerful combination of cryo-electron microscopy and tomography to generate high-resolution images of the earliest steps in the BK virus life cycle.
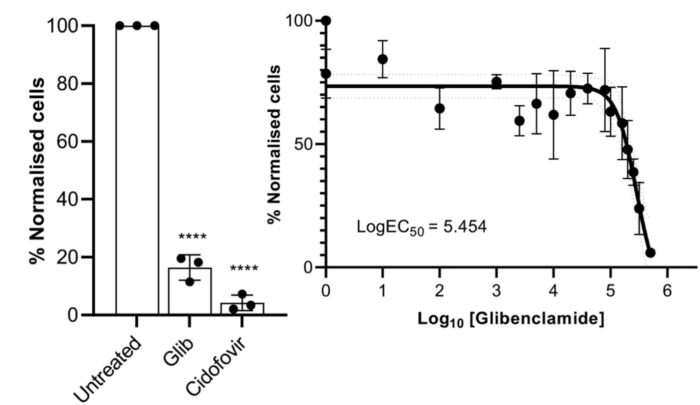
Targeted therapeutics for virus infection.
As obligate intracellular parasites viruses must utilise the resources of the infected host cell in order to replicate and release new virus progeny. This dependence on host replication apparatus serves as a potential Achilles Heel in that effective pharmacological modulation of the host factor has the potential to impair virus replication. We use large scale screens of inhibitor compounds to identify new targets to block virus replication. Through these studies we have shown a dependence of both HPV and human polyomaviruses for host cell ion channels.