Dr Antreas Kalli
- Position
- University Academic Fellow
- Areas of expertise
- molecular dynamics simulations; molecular modelling; membrane proteins; lipid membranes; ion channels
- a.kalli@leeds.ac.uk
- Location
- 6.30 LIGHT Building
- Faculty
- Medicine and Health
- School
- Medicine
Introduction
Membrane proteins play a critical role in many biological processes. They regulate the transmission of signals across cell membranes, help cells sense and respond to stress, and regulate how molecules move in and out of cells. Malfunction of membrane proteins can lead to a number of diseases, like diabetes and cardiovascular diseases.
Computational techniques, such as molecular dynamics simulations, enable us to follow the dynamics of membrane proteins in the membrane environment in which they function. Therefore, molecular dynamics simulations can provide structural and dynamic details about protein complexes and biological membranes in exquisite molecular detail.
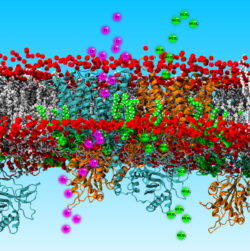
My group uses molecular dynamics simulations at different resolutions (i.e. coarse-grained and all-atom) to provide molecular/structural, and dynamic details about membrane proteins and membranes. Our research has a particular focus on constructing complex asymmetric model membranes that resemble native cell membranes in order to study the conformational dynamics of membrane proteins in more realistic lipid environments, like the ones in which these proteins function. By using such simulations we study how proteins interact with their lipid environment and with partner proteins.
Current major projects
- Understanding the function of Piezo1 and other mechanosensitive ion channels.
- Investigating the function and interactions of membrane transport proteins.
- Understanding the interaction of peripheral membrane proteins with lipids.
- Studying the activation mechanism of signalling receptors
Detailed research programme
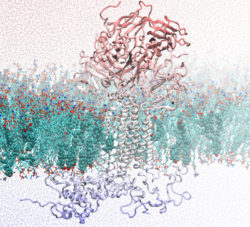
Understanding how the Piezo1 mechanosensitive channel is regulated and activated.
Mechanical forces regulate many vascular functions including endothelial cell alignment to blood flow, and cardiac remodelling. Our aim is to examine how forces are sensed in the cardiovascular system. Our research focuses on the Piezo1 channel. Piezo1 is a critical mechanical sensor in endothelial cells, red blood cells, and other cell types such as cardiac fibroblasts. In this project, we use molecular dynamics simulations and molecular modelling to examine how the Piezo1 channel transitions between active and inactive states, how it senses mechanical force, and how it transports ions. We also use computational methodologies to study how disease-causing mutations may alter Piezo1 activation.
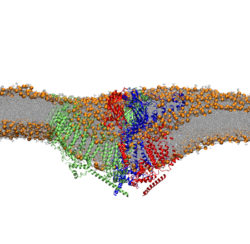 Elucidating the interactions and transport mechanism of the human chloride/bicarbonate anion exchanger 1
Elucidating the interactions and transport mechanism of the human chloride/bicarbonate anion exchanger 1
Anion exchanger 1 (AE1, Band 3), a member of the Solute Carrier 4 family of transporters, is found in red blood cells, where it functions as an electroneutral chloride/bicarbonate exchanger. This process is crucial for efficient respiration. Mutations in AE1 lead to inherited diseases such as Southeast Asian Ovalocytosis (SAO) and anaemias. In this project we use molecular dynamics simulations and molecular modelling to study the AE1 transport mechanism and its interaction with membrane lipids. We also use large-scale molecular dynamics simulations to study the large-scale organization of AE1 in model membranes that mimic the red blood cell plasma membrane.
Elucidating the interactions of peripheral membrane proteins with lipids.
The binding of peripheral membrane proteins to cell membranes is regulated by lipid-binding modules that are part of these proteins. These lipid binding domains e.g. PH, PX, SH2, and FYVE often associate with specific lipids in membranes e.g. phosphatidylinositol (PIP) lipids. We use coarse-grained and atomistic molecular dynamics simulations to study the mechanistic details of the binding, interactions and diffusivity of lipid-binding domains to cell membranes. We also develop high-throughput computational approaches that allows us to study proteins faster and more efficiently. These approaches enable us to understand how large sets of lipid binding domains of the same family of proteins – as opposed to individual domains - attach to cell membranes.
Studying the activation of the T-cell receptor
The T-cell receptor (TCR) is a membrane protein located in T-lymphocytes. It is critical for the survival and function of T-cells and for the immune response. We use molecular dynamics simulations in which we simulate the TCR and related proteins e.g. lck in complex model membranes that resemble the native T-lymphocytes membrane to investigate: (i) the activation of TCR, and (ii) its interactions with cytosolic proteins and with lipid molecules at the molecular level. These studies provide a molecular understanding of the activation of the TCR. We use biomolecular simulation protocols developed for TCR to also study other signalling receptors, i.e. integrins.