Dr Charlie Scarff
- Position
- British Heart Foundation Jacqueline Murray Coomber Fellow & University Academic Fellow in Structural Biology
- Areas of expertise
- Integrative Structural Biology, Cryo-electron Microscopy, Mass Spectrometry, Muscle & Myosin, Cardiovascular Disease
- Location
- Level 7 LIGHT Laboratories
- Faculty
- Medicine and Health
- School
- Leeds Institute of Cardiovascular and Metabolic Medicine, School of Medicine
Introduction
My lab is interested in understanding structural-functional relationships in muscle and how defects in muscle proteins lead to cardiovascular disease. We use an integrative structural biology approach to do this, primarily combining cryo-electron microscopy, mass spectrometry, and mutational analyses with modelling. We are also passionate about the development of integrative structural biology methods for the study of cardiovascular disease as technology drives research. Our current focus is on studying how mutations in two muscle proteins, beta-cardiac myosin and myosin-binding protein C, lead to the inherited heart disease hypertrophic cardiomyopathy. We also have an interest in dilated cardiomyopathy, diabetic cardiomyopathy and the effects of glycation on muscle proteins, and structure-based drug design. This research will lead to a better understanding of the molecular mechanisms that underpin heart disease, and it is our hope that this will pave the way to better therapies in the future.
Current major projects
- The structural basis of inherited heart disease
- The interacting-heads motif of myosin
- Cardiac myosin-binding protein C structure and function
- Integrative structural biology
Detailed research programme
The structural basis of inherited heart disease
Hypertrophic cardiomyopathy (HCM) is an inherited heart disease, which affects between ~0.2-0.5 % of the population and is the most common cause of heart failure in the young. Most people with the disease have mutations in either beta-cardiac myosin heavy chain or myosin-binding protein C but it is not known how these mutations lead to disease. Myosin, the molecular motor responsible for muscle contraction, is able to be switched off between contractions to regulate activity and conserve energy. My group uses a structural approach to explore whether disease-causing mutations affect myosin’s ability to switch off, leading to increased energy use and cardiac remodelling.
The interacting-heads motif of myosin
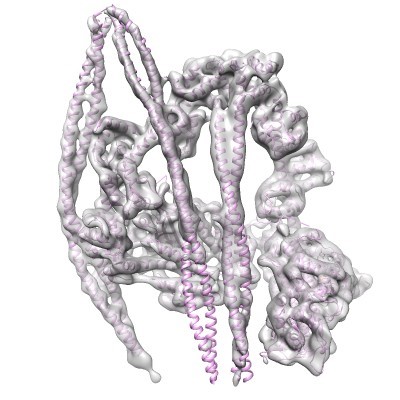
The interacting-heads motif (IHM) of myosin is a switched-off energy-conserving state. It is proposed that hypertrophic cardiomyopathy mutations could destabilise this state. Currently, we lack a high-resolution structure of beta-cardiac myosin in the IHM to elucidate the effect of mutations on its structure. In collaboration, we have recently solved the structure of the shutdown state of smooth muscle myosin (shown in the picture, Scarff et al., Nature, 2020) and this now serves as a model for the beta-cardiac myosin IHM. This is allowing us to explore the effects of mutation and small molecules on this structure and raises the prospect of structure-based drug design.
Cardiac myosin-binding protein C structure and function
Cardiac myosin-binding protein C (cMyBP-C) is a major protein of muscle thick (myosin) filaments with structural and regulatory roles in the heart. We lack a clear understanding of how cMyBP-C functions, and what its molecular interactions are within the muscle contractile unit (sarcomere). Mutations in cMyBP-C are responsible for 25 % of HCM disease cases. We seek to understand the structure and function of cMyBP-C, the interactions cMyBP-C makes within the sarcomere, and the effects of disease relevant mutations.
Integrative structural biology
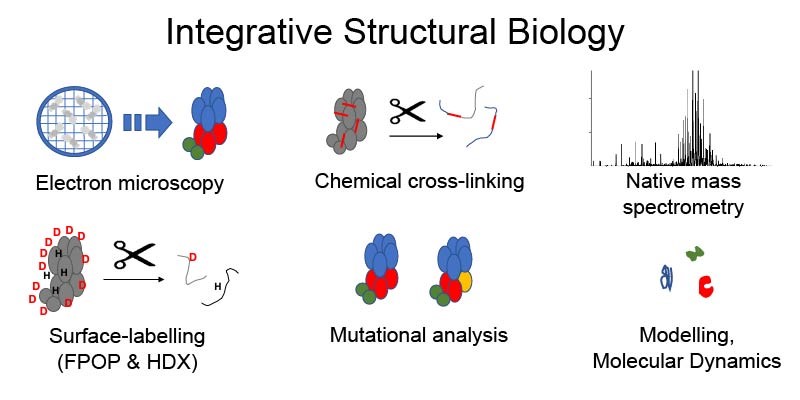
My lab is interested in how electron microscopy-based workflows can be combined with mass spectrometry-based workflows to enable increased understanding of macromolecular complexes, their dynamics and heterogeneity. The image demonstrates an integrative structural biology approach, combining electron microscopy, native mass spectrometry, surface labelling, chemical cross-linking, mutational analysis, and modelling workflows.