Dr Darren Tomlinson
- Position
- Associate Professor
- Areas of expertise
- Affimers; protein interactions; structural biology; cancer; drug discovery
- Phone
- +44(0)113 343 7099
- Location
- 7.108 Astbury
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
- Website
- Google scholar
 Introduction
Introduction
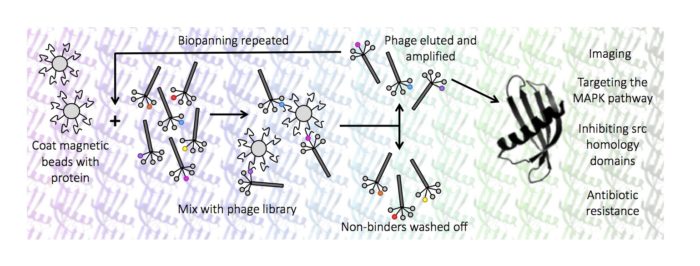
Our research group is interested in using a novel molecular recognition tool, called an Affimer, to study protein function. An Affimer (also known as an Adhiron) is a scaffold protein that constrains two randomised loops for molecular recognition. They can be used as alternative reagents to antibodies for applications like diagnostics and used to probe the surface of target proteins to identify key domains involved in protein function. We use a range of techniques from molecular biology, phage display, structural biology, cellular biology and chemical biology to understand proteins in molecular detail. We study a range of proteins and protein families involved in normal and disease processes.
Current major projects
- Targeting the MAPK pathway
- Inhibition of src homology domains
- Targeting antibiotic resistance
- Developing methods for imaging and super-resolution microscopy
Detailed research programme
Targeting the MAPK pathway
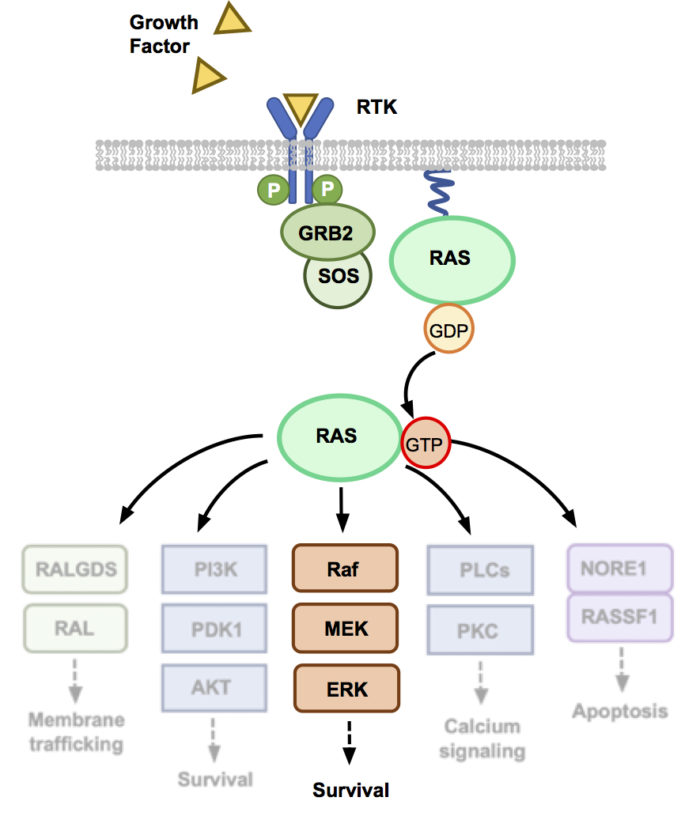
Our work on the MAPK pathway uses Affimers to target and understand proteins involved in the MAPK pathway, including RAS, SOS and PDKs. Mutations in RAS are present in ~25% of all cancers but, until recently, few therapeutics have successfully reached the clinic. In addition, the role of SOS in mutant RAS driven cancers has yet to be fully explored. We demonstrate a range of disciplines from structural biology using X-ray crystallography, to intracellular proximity-based assays such as NanoBRET, to fully understand the effect of our protein binders on this oncogenic pathway. The use of intracellularly expressed Affimers allows us to understand the impact of inhibiting different proteins in the MAPK pathway and the survival of cancer cell lines with different genotypes. As Affimers can probe hotspots on the surface of challenging proteins, we also explore their use as a template pharmacophore for small-molecule identification.
Inhibition of src homology domains
Protein interaction domains play a critical role in regulating the specificity of signal transduction, through their ability to recognise and recruit specific targets. Examples of these domains include Src homology 2 (SH2) and Src homology 3 (SH3). SH2 domains typically bind phosphorylated tyrosine residues on proteins that mediate receptor tyrosine kinase signalling. SH3 domains bind proline-rich peptides and mediate protein-protein interactions (PPIs) of signalling proteins, such as Grb2, and proteins of the cytoskeleton.
Aberrant PPIs contribute to human diseases, such as cancer. However, study of PPIs is hampered by the lack of specific reagents targeting these interactions. In the Tomlinson group, we have isolated and characterised Affimer reagents that specifically inhibit interactions with SH2 and SH3 domains. We are using these reagents in cellular and structural studies to understand their role in signalling pathways and to help inform drug discovery and chemical biology approaches to targeting conserved proteins.
Targeting antibiotic resistance
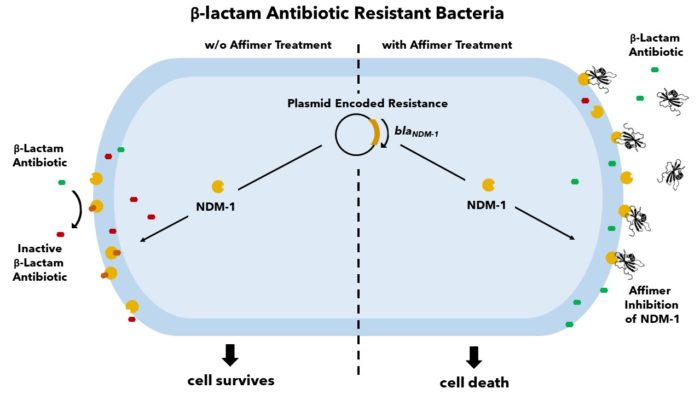
The increasing prevalence of antibiotic-resistant bacterial infections is of urgent global concern. New Delhi Metallo-β-lactamase 1 (NDM-1) is a carbapenemase capable of hydrolysing the β-lactam ring in almost all known β-lactam antibiotics, thus conferring broad range antibiotic resistance. With few antibiotics effective on NDM-1 expressing bacteria and resistance quickly developing to our last resort therapies, it is crucial to develop new treatments. Affimers have been identified to bind and inhibit NDM-1 in a dose-dependent manner and extensive in vitro inhibition characterisation has revealed detailed binding specifics. Through cellular assays, we have verified that antibiotic-resistant NDM-1 expressing strains regain antibiotic susceptibility upon treatment with Affimers. We are exploring the use of these Affimers in the development of novel NDM-1 inhibitors as well as exploring the potential of targeting other similar proteins to combat this disease.
Developing methods for imaging and super-resolution microscopy
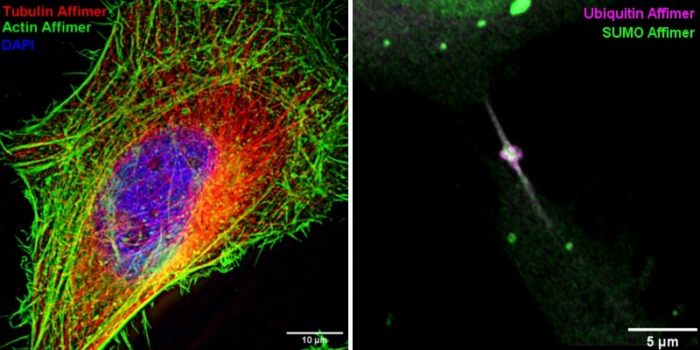
The specificity and affinity of Affimers make them an effective alternative to antibodies for use in histochemistry and fluorescence microscopy. Their small size facilitates improved access to epitopes and reduces ‘linkage error’ which is especially advantageous for super-resolution techniques. To further reduce linkage error, we are developing strategies to conjugate organic dyes directly onto proteins of interest via Affimer-directed fluorophore warhead delivery. Expression of Affimers fused to fluorescent proteins or self-labelling tags enables imaging of endogenous proteins within living cells, of use for example to study dynamic processes such as cell division. We are using Affimers to image a wide range of targets including cytoskeletal components (actin, tubulin), cell surface receptors (HER4, VEGFR2) and SUMOylated and ubiquitinated proteins.