Dr Jenny Tomlinson
- Position
- Royal Society Dorothy Hodgkin Research Fellow
- Areas of expertise
- NMR spectroscopy, antimicrobial resistance, protein dynamics, PPIs, structural biology
- Location
- 6.02 Miall Building
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
- Website
- ORCID
Introduction
Antimicrobial resistance is a major public health concern and has been predicted to overtake cancer as a cause of death by 2050. Understanding the molecular detail of antibiotic resistance mechanisms raises the possibility of attempting to overcome them and so rejuvenate the use of clinically important antibiotics. The roll of allostery and conformational dynamics in antibiotic resistance is not well understood but recent studies have identified mechanisms in which they play a key role. My research uses NMR spectroscopy combined with other biophysical and biochemical techniques to understand these mechanisms in molecular detail.
Current major projects include:
- Elucidating the mechanism of FusB mediated fusidic acid resistance.
- Understanding the allosteric control of PBP2a activation.
- Inhibition of the FusB:EF-G protein:protein interaction.
Detailed research programme
Understanding FusB-mediated fusidic acid resistance
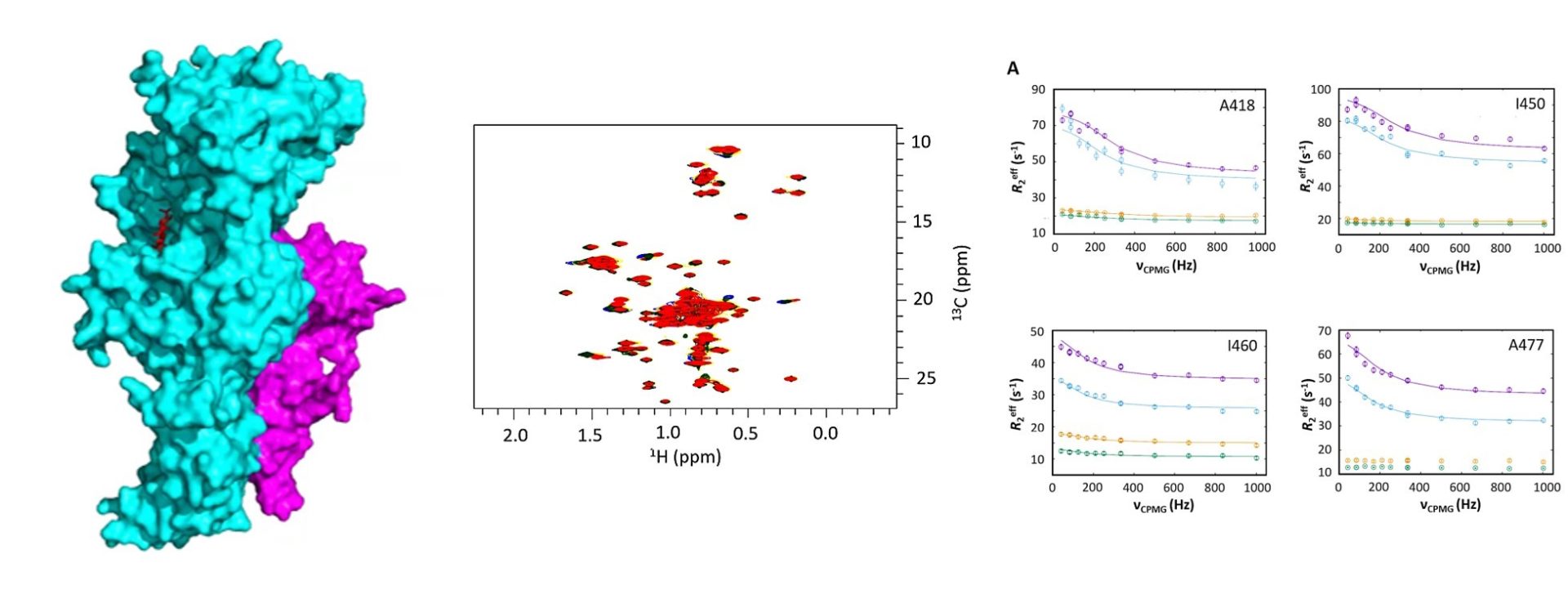
Fusidic acid is a clinically important antibiotic that can be used to treat MRSA but resistance has increased in recent years, driven by expression of the FusB family of proteins. FusB rescues the drug target (EF-G) from inhibition by fusidic acid through binding to EF-G and promoting dissociation of stalled EF-G/ribosome/fusidic acid complexes. Our recent research using NMR relaxation dispersion and biochemical assays has shown that an allosterically induced change in the dynamics of EF-G is important in driving this resistance mechanism. We are now working to fully elucidate the molecular details of this resistance mechanism and understand the allosteric control within EF-G.
Understanding the allosteric control of PBP2a activation
PBP2a confers resistance beta lactam antibiotics by providing an alternative PBP protein for peptidoglycan synthesis that has a closed active site inaccessible to antibiotics. Peptidogylcan precursors open the active site by binding to a distant allosteric site and promoting conformational change. We are studying the transmission of this conformational change though the protein to understand the mechanism of methicillin resistance.
Inhibition of the FusB:EF-G protein:protein interaction
To overcome FusB-mediated resistance to fusidic acid, we are working to understand the details of the FusB-EF-G protein-protein interaction with a view to inhibiting this interaction to prevent resistance to fusidic acid. We are studying the effects of single amino acid substitutions on the affinity of the interaction to identify key residues and designing peptides to try to inhibit the interaction.