Dr Joan Boyes
- Position
- Associate Professor
- Areas of expertise
- Gene regulation, Epigenetics, V(D)J recombination, Leukaemia, Antibody diversity
- j.boyes2@leeds.ac.uk
- Location
- Astbury 10.103
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
- Website
- ORCID
Introduction
V(D)J recombination generates a highly diverse repertoire of immunoglobulin and T cell receptor genes that allow an almost infinite number of potential pathogens to be combatted. However, the recombination reaction involves the breakage and rejoining of DNA and is inherently risky. Consequently, the price of having such an effective immune system is genome instability and cancer. We are investigating the mechanism by which V(D)J recombination is regulated and how mistakes in this reaction can lead to oncogene activation.
Current major projects:
- Epigenetic regulation of V(D)J recombination
- Regulation of end joining during V(D)J recombination
- Determining the impact of reintegration of the ESC
- Analysis of the aberrant recombination reaction, cut-and-run
Detailed research projects
Epigenetic regulation of V(D)J recombination
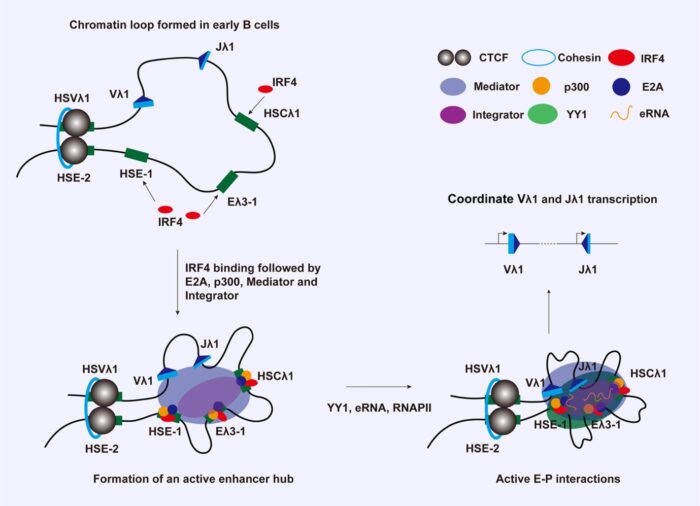
Epigenetic modifications are central to restricting V(D)J recombination to the correct cell type and correct stage of development. We are investigating which chromatin changes are required using the mouse immunoglobulin lambda light chain locus (Igλ) as a model. This is the smallest antigen receptor locus with only six functional recombining gene segments. The Igλ locus rearranges primarily in pre-B cells but we showed that increased levels just of a single transcription factor at the earlier pro-B cell stage are enough to completely activate the Igλ locus. Capitalising on this, we developed a system in which Igλ recombination can be induced. This allows us to follow the steps during recombination temporally via examining the order of regulator binding, changes in local epigenetic marks as well as changes in long range chromatin interactions.
Regulation of end joining during V(D)J recombination
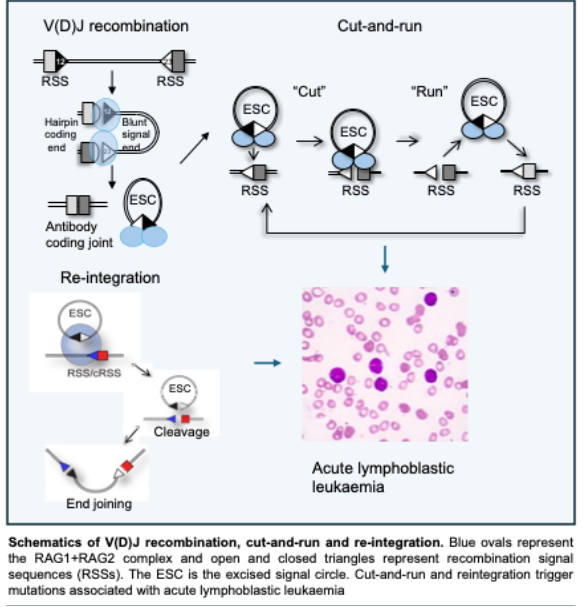
During V(D)J recombination, two gene segments, flanked by recombination signal sequences (RSSs), are brought together into a synaptic complex. V(D)J recombinase cutting generates four broken DNA ends: Two at the coding gene segments and two ends that terminate in the RSSs (signal ends). It is essential that the coding segments are joined together to generate the variable exon of the immunoglobulin or T cell receptor. The signal ends are usually joined into a circle that is excised from the genome as a by-product, the excised signal circle (ESC). We are investigating how the RAG recombinase shepherd the coding ends into the correct end joining complex to ensure efficient production of antigen receptor genes.
Determining the impact of reintegration of the ESC
The ESC by-product of V(D)J recombination retains the RSSs. This means that the recombinase can re-bind to the ESC. The resulting recombinase/ESC complex then targets RSSs or RSS-like sequences (known as cryptic RSSs or cRSSs) in the genome, resulting in reintegrate the ESC. The danger posed by reintegration depends on its location. For example, reintegration of large amounts of DNA (up to 1Mb) from the ESC into a tumour suppressor gene will contribute to cancer development. We are investigating the influence of ESC reintegration in B-cell acute lymphoblastic leukaemia (B-ALL) using bespoke programmes and whole genome sequences of B-ALL patients.
Analysis of the aberrant recombination reaction, cut-and-run
The ESC by-product is involved in a second, related reaction, known as cut-and-run. Here, the recombinase/ESC complex binds to a RSS or cRSS (just like reintegration). However, in this case, only the RSS/cRSS is cut. The recombinase/ESC complex dissociates, leaving a double strand break. Remarkably, breaks that have been caused by cut-and-run map to many of the frequently mutated genes in B-ALL and furthermore to the genes that are frequently mutated at relapse. This suggests that cut-and-run underpins many key mutations involved in disease progression. We are exploring the fundamental mechanisms of cut-and-run to identify ways to suppress this reaction.