Dr John Barr
- Position
- Associate Professor
- Areas of expertise
- Structure, function and cell biology of negative-sense RNA viruses
- j.n.barr@leeds.ac.uk
- Location
- Astbury 8.108
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
Introduction
The group of negative sense RNA viruses include serious pathogens of plants, animals and humans, with many classified as ‘emerging viruses’ due to their appearance in new ecological niches. Within this broad grouping, we study members of both the family Paramyxoviridae and the order Bunyavirales, with an aim to understand how these viruses are built from their structural components, as well as how they hijack native host cell processes to aid in their replication. Our work is multidisciplinary, involving structural biology, high-resolution light and electron microscopy, as well as molecular virology and cell biology techniques. Our overarching goal is to identify interactions that are critical to virus replication cycles that may represent targets for intervention, as well as use viruses as tools to better understand native host cell processes.
Current major projects
- Virus ribonucleoprotein structure and function
- Visualising virus factories
- Host cell/pathogen interactions
- Ionic triggers for virus infection
Detailed Research programme
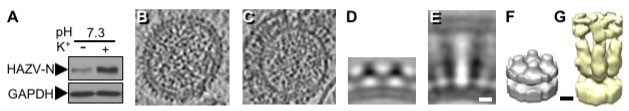
Ionic triggers for virus entry
Enveloped viruses express fusion protein spikes that promote merging of viral and cellular membranes, allowing entry of viral genetic material into cells. These spikes are activated by a variety of stimuli that ensure fusion only occurs in appropriate cellular locations, thus increasing virus infectivity. We recently revealed that potassium ions are a trigger for spike fusogenesis. Using both Bunyamwera and Hazara viruses, model members of the Bunyavirales order, we showed that certain potassium ion concentrations are reached within specific compartments of the endocytic network, and these act as biochemical triggers promoting spike conformational changes and activation of the fusion process. We aim to identify other ionic fusion triggers for other viruses, and also understand the processes that lead to ionic accumulation within membranous cellular compartments.
Virus factories
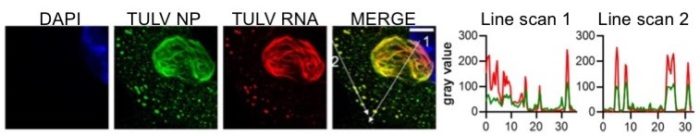
One of the many ways in which viruses hijack cellular factors and processes is in rearranging intracellular architecture to establish sites where viral components are synthesised for assembly into new virions. These sites are known as ‘virus factories’ and they often are situated within various endo-membranous compartments. We use both light and electron microscopy to identify and characterise these sites, to better understand how viruses recruit cellular components and induce radical reorganization of the cell interior.
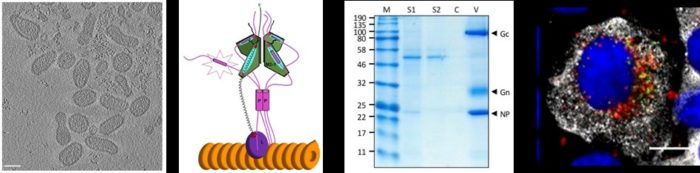
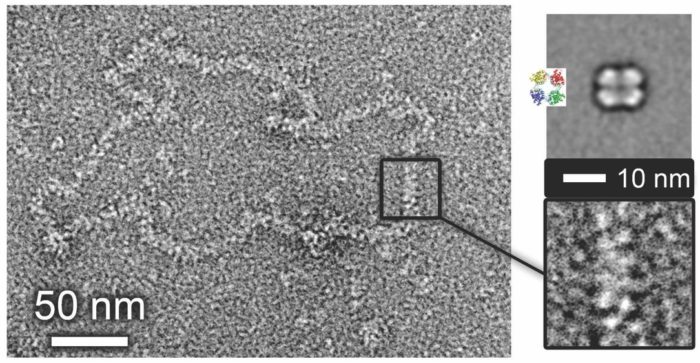
Virus ribonucleoprotein structure and function
Negative sense RNA viruses enwrap their genomes with a virus-encoded RNA binding protein called the nucleocapsid protein (NP). Multiple copies of the NP associate with the genome to form a large assembly called a ribonucleoprotein (RNP) complex, and assembly of this RNP is essential for many aspects of the viral multiplication cycle from gene expression to virion assembly. Using high-resolution structural techniques available within the Astbury centre, combined with reconstitution of RNPs in cells using reverse genetics, we aim to derive a detailed understanding of the RNP structure/function relationship for many candidate viruses within the bunyavirus and paramyxovirus groups.