Dr Megan Wright
- Position
- Associate Professor
- Areas of expertise
- chemical biology; proteomics; chemical probe; host-microbe; organic synthesis
- Phone
- +44(0)113 343 3196
- Location
- G.18b Chemistry
- Faculty
- Engineering and Physical Sciences
- School
- Chemistry
Introduction
Our group works in the field of chemical biology, applying chemical tools to answer questions about dynamic protein function and the mode of action of small molecules in live cells and at the molecular level. This research is highly interdisciplinary and spans organic and peptide synthesis, protein biochemistry, cell biology and quantitative mass spectrometry-based proteomics. We work closely with scientists from other disciplines, enabling us to design innovative tools for cell biology and to tackle a wide range of biological questions – from the mechanisms of antibacterial resistance, to how plants regulate growth, and cancer cell signalling pathways.
Current major projects
- Identifying the targets of bioactive small molecules in live cells
- Chemical and structural approaches to protein-protein and protein-ligand interactions
- Chemical tools to understand the redox regulation of proteins
Detailed research programme
Target identification in cells
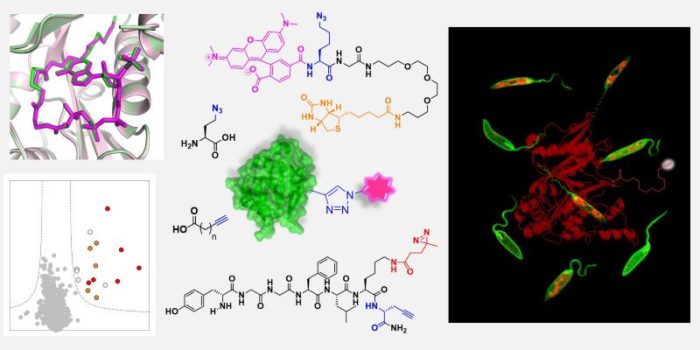
Identifying the protein targets of molecules in live cells is very challenging in such a complex biological environment. One area of our research is in equipping small molecules with photo- or chemically-reactive functionalities to stabilise their interactions with protein targets. These probes also contain small, minimally disruptive tags for subsequent selective labelling via click chemistry. This allows us to detect, isolate and identify protein interactors. Projects in this area include identifying the targets of anti-parasitic compounds, and of small molecules and peptides that mediate communication between bacteria and their hosts.
Mapping and imaging protein-protein and protein-ligand interactions
We are also applying protein-reactive chemistry to trap and analyse dynamic protein-protein and protein-ligand interactions. We are designing chemical tools mimicking bacterial peptides that are recognised by receptors on the surface of human cells, for example during the immune response to infection. These tools are allowing us to detect this protein-ligand interaction with high sensitivity and to identify the binding site. In another project, we are applying a suite of reactive crosslinkers to trap dynamic protein complexes in distinct conformations; advanced mass spectrometry approaches allow us to map and then model these complexes.
Chemical tools to understand the redox regulation of proteins
We are developing chemical tools and methods to map reactive protein cysteines in cells with spatial and temporal resolution. These tools will enable us to study redox (reduction-oxidation) signalling, where information flows through a biological system by the reaction of small molecules with protein thiols. Redox signalling is a crucial mechanism by which cells respond to their environment and coordinate internal processes such as cell division. Cells tightly control redox status across subcellular organelles, but we currently lack the tools to study, across the proteome, where the proteins that sense redox status are localised.