Dr Morgan Herod
- Position
- MRC CDA Fellow and University Academic Fellow
- Areas of expertise
- RNA viruses; Virus Replication; Virion Morphology; Virus-Host Interactions; Viral Polyproteins
- Location
- 10.27 Miall
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
Introduction
The Herod lab has a broad interest in virus replication, with a particular interest in studying single-stranded positive-sense RNA viruses important for human and animal health. We uses an interdisciplinary approach to understand the molecular mechanism that regulate viral genome replication, virion assembly and disassembly. The lab studies a range of viruses, with a particular interest in GI viral infections such as norovirus and hepatitis E virus. Out overarching aim is to use a deeper understanding of the molecular mechanisms of viral replication to develop new approaches to disease control.
Current major projects
- How do viral capsids disassemble and enter the cell?
- How does viral polyprotein processing control replication?
- Which molecular interactions control virus genome replication?
- How are new virions assembled?
Detailed research programme
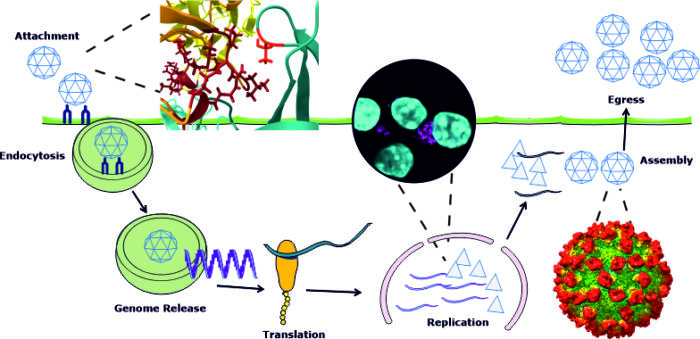
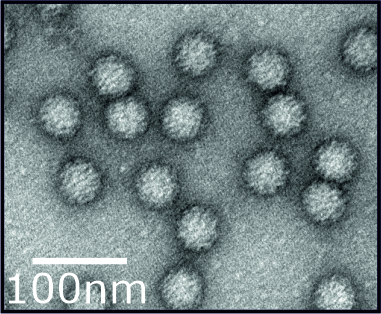
How does the norovirus capsid interact with cells and molecules to disassemble and regulate entry into the cell?
Norovirus particles consist of a genome enclosed with a protein shell or capsid. These capsids are important for protecting the genome and transmitting it between hosts. We use a broad range of techniques spanning molecular and structural biology, classical virology and genetics to understand the importance of the norovirus capsid during the early part of the virus life-cycle. This includes how the capsids interacts with ligands and biomolecules, the conformational changes that can occur to capsids and how these factors control endocytosis, virion structure and infectivity. These are all essential viral processes that are required to for examples release the genome to initiate an infection.
Which molecular interactions control viral genome replication and assembly of new virions?
In the latter part of a viral life-cycle new virus particles are assembled from newly synthesised genomes and capsids proteins. Synthesis of new viral genomes is thought to occur with virally produced organelles sometimes referred to as “replication complexes”. A complex network of interactions controls how viral genomes synthesised within these complexes are assembled into new virions. Using structural, molecular and cell based techniques we are dissecting the molecular composition of viral replication complexes, in particular norovirus and picornavirus replication organelles, how new virions are assembled from these complexes.
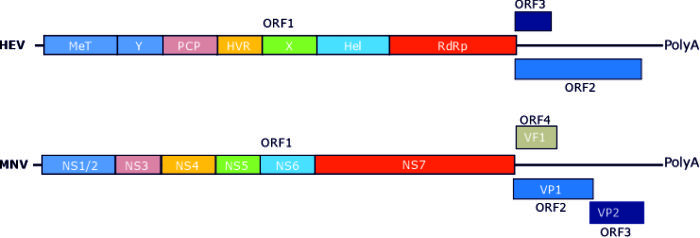
How does processing of the hepatitis E virus polyprotein regulate viral genome replication?
Hepatitis E virus is a leading cause of acute viral hepatitis in the UK and Europe with a substantial mortality rate in certain populations (e.g. during pregnancy). Viral RNA replication is performed by virus encoded non-structural proteins that are produced via a polyprotein. We used molecular virology systems to reveal how this viral polyprotein is processed to generate functional non-structural proteins, the importance of this processing in regulating replication and ultimate the functions of the individual non-structural proteins in the viral “replication complex”.