Dr Roman Tuma
- Position
- Visiting member
- R.Tuma@leeds.ac.uk
Introduction
We apply combination of advanced biophysical and single molecule imaging techniques to study self-assembly and function of large macromolecular complexes. We study assembly and genome packaging of complex viruses from the Reoviridae family and the role of liquid-liquid phase separation in the formation of virus replication factories. We are linking intrinsic dynamics of Sec machinery to its function during ATP-driven translocation of polypeptide chains across cellular membranes.
Current major projects
- Replication factories of Avian Reovirus - Assembly and nucleic acid packaging in dsRNA viruses
- Mechanism of ATP-driven polypeptide translocation across membranes- Sec translocon dynamics
- Single molecule fluorescence – dynamic properties of large assemblies
Detailed research programme
Replication factories of Avian Reovirus and packaging of segmented dsRNA genomes
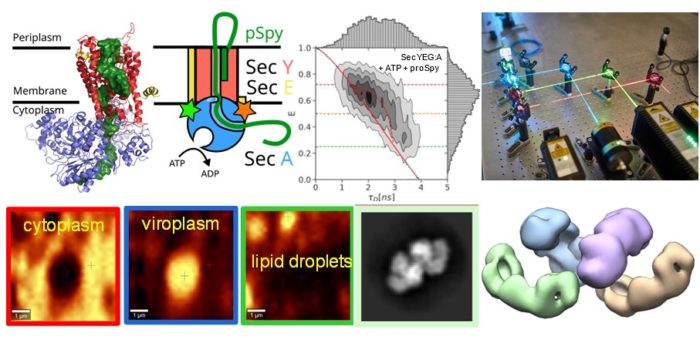
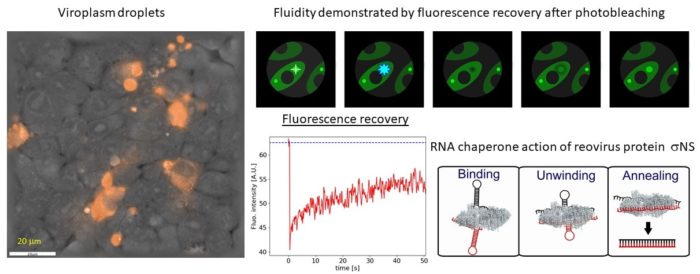
Viruses belonging to the Reoviridae family exhibit segmented dsRNA genomes and replicate and assemble in so-called virus factories (also known as viral inclusion bodies or viroplasms). Using various spectroscopic (Raman and fluorescence recovery after photobleaching) and holographic refractive index imaging techniques we demonstrate that viroplasms are dense membrane-less compartments that are formed by liquid-liquid phase separation in the cytoplasm. Virally encoded RNA binding proteins, such as avian reovirus σNS, are major constituents of viroplasms. We demonstrate that σNS forms hexamers and octamers in solution. The octamer is effective in unfolding local RNA secondary structure and binds two RNA strands simultaneously, thus facilitating formation of stable inter-segment RNA-RNA interactions. Such interactions were implicated in the genome assortment and RNA packaging. On longer RNA substrates σNS readily forms helical filaments which may protect RNA prior to packaging and facilitate search for complementarity among viral segment precursors. Thus, σNS is a versatile RNA chaperone with RNA binding properties that are optimized for remodelling of local RNA structure while facilitating search for sequence complementarity between segment precursors.
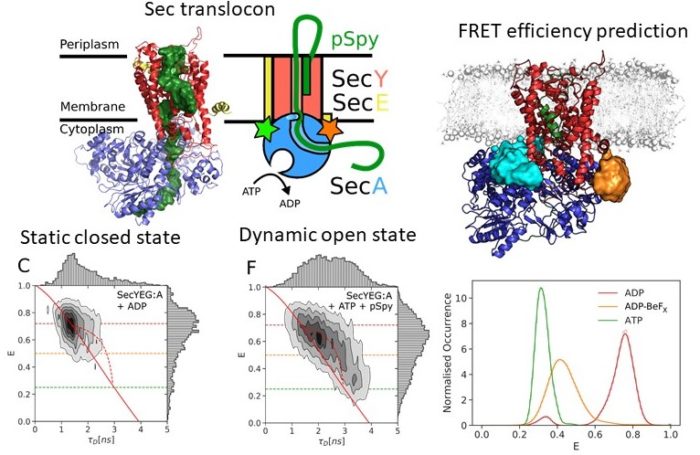
Mechanism of ATP-driven polypeptide translocation across membranes by the Sec translocon
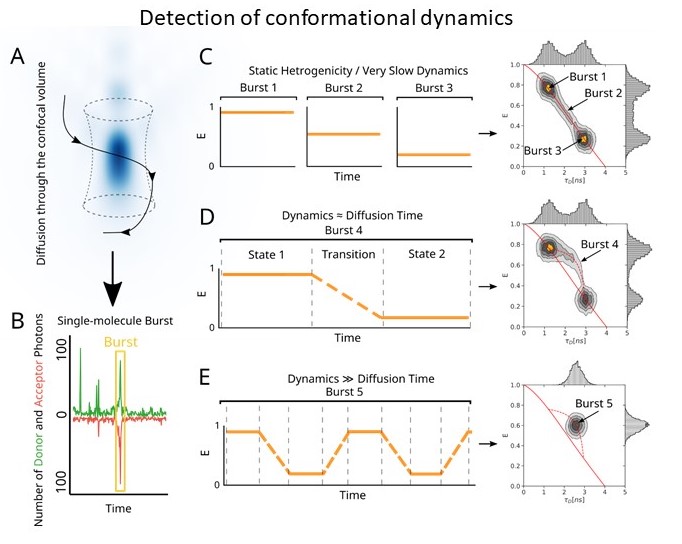
The Sec translocon is conserved among all forms of life as the principal route for the efficient transport of polypeptides across membranes and for insertion into lipid bilayers. In bacteria, translocation of unfolded polypeptides into the periplasm is primarily driven by concerted action of SecA ATPase and the SecYEG integral membrane complex. ATP binding to SecA promotes opening of SecY lateral gate (LG) while ADP closes the gate and this in turn modulates transport of polypeptide chain through the central pore. Here we set to elucidate the nature of the allosteric communication between SecA ATP binding site and SecYEG LG using time resolved single molecule fluorescence. Direct observation of LG transitions reveal that slow conformational changes associated with SecA ATPase (~ 6 s-1) modulate fast LG motions (~ 175 s-1). We propose a new dynamic allosteric mechanism in which SecA ATPase cycle steers energy landscape that in turn governs SecY LG motion. Analysis of molecular dynamic trajectories further pinpointed key molecular interactions involved in the mechanism. The mismatch in the rates of ATP hydrolysis and lateral gate opening is consistent with SecYEG acting as a Brownian-ratchet.
Single molecule fluorescence
We widely collaborate on projects involving single molecule fluorescence. Our lab is equipped with home built total internal reflection microscope suitable for smFRET with time resolution 50 ms per frame. This is complemented by a pulsed interleaved confocal setup equipped with nanosecond time resolved detection. This offers improved time resolution (0.1-50 ms time scale) and allows investigation of dynamic interconversion between conformational states. We are developing these techniques further at our new location (http://makrokomplex.cz/facilities/single-molecule-fluorescence-microscopy/)