Dr Stephen Muench (Deputy Director)
- Position
- Assistant Professor in membrane proteins
- Areas of expertise
- structural biology; electron microscopy; membrane proteins; time-resolved
- Phone
- +44(0)113 343 4279
- Location
- 6.106 Astbury
- Faculty
- Biological Sciences
- School
- Biomedical Sciences
- Website
- Google scholar
Introduction
Structural biology has often helped support our ability to generate new therapies through furthering our fundamental understanding of cellular processes and structure-based drug design. However, structural biology still has a number of challenges, not least in our ability to study membrane protein systems and to trap short lived (ms) intermediates such as domain re-organisation upon substrate binding. Our lab is developing new tools to permit time-resolved methodologies for electron microscopy that can trap states in the low ms time-frame (>5ms). Moreover, we are studying new approaches to stabilising membrane proteins and investigating how the closely associated lipids may play a role in regulation, function and stability. This allows us to better understand the protein targets we are studying which can inform our structure-based drug design programs.
Current major projects
- Structural and mechanical studies of membrane proteins
- Using electron microscopy to drive small molecule development
- Developing time-resolved applications for electron microscopy
- Developing new tools for membrane protein extraction and stabilisation
Detailed research programme
Small molecule design
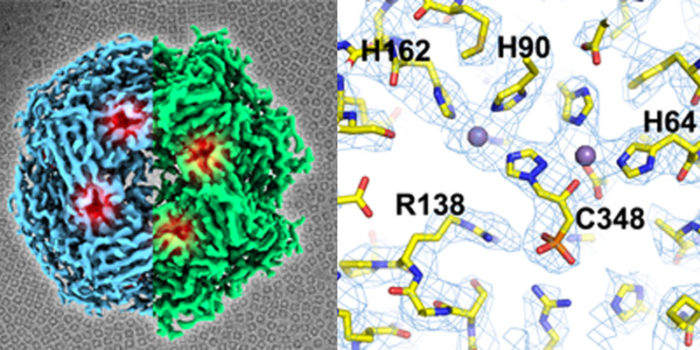
Traditionally techniques such as X-ray crystallography have underpinned structure-based drug design. However, in recent years there have been a growing number of examples showing the use of single particle cryo-EM to inform on small molecule binding. We used single particle cryo-EM to reveal the difference in sensitivity between the yeast and plant homologues of IGPD and have used cryo-EM to study inhibitor binding to the cytochrome bc1 complex amongst other systems. We have a number of active studies now combining the structural insights from our cryo-EM experiments and high throughput docking to develop treatments to disease including toxoplasmosis, epilepsy, cancer and heart disease.
Time-resolved methodologies
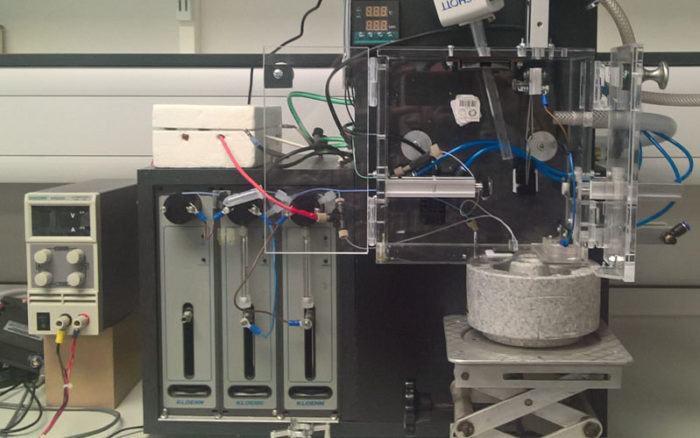
We have been developing new approaches to grid making for single particle cryo-EM with a focus on the rapid mixing, spraying and plunge freezing of grids. The current apparatus allows us to trigger and stop reactions >5ms to study for example, virus disassembly, motor protein function, protein polymerisation and the cycling of large protein complexes. Moreover, this has allowed us to investigate how the speed of grid preparation can also play a significant role in the particle orientation and damage at the air-water interface. We are now looking at new approaches for delivering protein on a grid that can further improve the quality of the frozen sample on the grid and increase the time-frame in which we can work.
Investigating the role of lipids in function and stability of membrane proteins
Lipids play a key role in membrane protein structure, function and regulation. However, with new tools available such as styrene maleic acid copolymers (SMA) we are now in a position to start to characterise the nature of these lipids and how they influence membrane protein function. Through the study of new polymers were developing a system that can work with biophysical approaches such as mass spectrometry and electron microscopy, providing insights not just on protein structure but also the closely associated lipid environment. For example, the group have been involved with a project to show how a potent small molecule works by replacing a lipid within the protein opening up new opportunities for targeting membrane protein function through lipid mimics.