Dr Yvonne Nyathi
- Position
- Lecturer in Membrane Biology
- Areas of expertise
- Membrane proteins, proteostasis, molecular chaperones, ribosomes, liquid-liquid phase separation
- y.nyathi@leeds.ac.uk
- Location
- Garstang 6.44e
- Faculty
- Biological Sciences
- School
- Biomedical Sciences
Introduction
Maintaining cellular protein homeostasis [proteostasis] is crucial for overall cell health and adaptation to stress. Disruptions in proteostasis, caused by factors like aging or genetic mutations, can lead to protein misfolding and formation of harmful aggregates. Around 25% of our genes encode membrane proteins which are vital for cellular function. However, around 5-10% of these proteins mislocalise to the cytosol due to defects in recognition or translocation. Once mislocalised, the hydrophobic nature of membrane proteins makes them prone to aggregation, contributing to cellular dysfunction and diseases like Alzheimer's. Our laboratory focuses on unravelling the molecular mechanisms governing membrane protein proteostasis, spanning synthesis to integration into target membranes. With over 50% of drug targets being membrane proteins, our research not only addresses fundamental cellular processes but also holds promise for therapeutic interventions. Employing a multidisciplinary approach, we integrate proteomics, high-resolution imaging, bioinformatics, biochemistry, and structural biology methods to advance our understanding of membrane protein function and dynamics.
Current major projects
- Dissecting the role of the SGTA/BAG6 complex in membrane protein quality control
- Elucidating the structure of the chaperone complex responsible for targeting and membrane protein triage.
- Identification of chemical modulators of membrane protein proteostasis
- Engineering membrane protein quality control systems for improved production of membrane proteins
Detailed research programme
Dissecting the role of the SGTA/BAG6 complex in membrane protein quality control
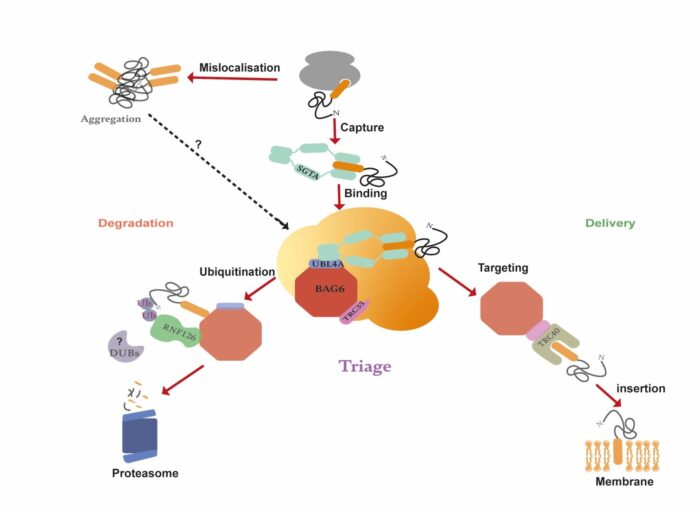
Our research focuses on unravelling the roles of the co-chaperone protein SGTA and its binding partner Bag6 in regulating the proteostasis of membrane proteins. The central goal of our work is to elucidate the precise molecular mechanisms underpinning the initial recognition and subsequent fate of mislocalised membrane proteins. Specifically, we are studying how the SGTA/BAG6 complex distinguishes between membrane proteins en-route to the endoplasmic reticulum (ER) and those mislocalised to the cytosol and how this dictates their fate. Simultaneously, we are using proteomic approaches to define the substrate specificity of the SGTA/BAG6 complex. Our work will lead to deeper understanding of membrane protein proteostasis and the identification of potential targets for therapeutic interventions in diseases characterised by mislocalisation or protein misfolding.
Elucidating the structure of the chaperone complex responsible for targeting and membrane protein triage
The Bag6/ SGTA complex has been implicated in cancer, neurodegenerative diseases, and viral infectivity. However, we still do not understand how this complex impact on these diseases. The absence of a structure for this complex remains a critical gap in our understanding. SGTA is a modular protein that engages with Bag6 through its N-terminus, interacts with Hsp70 and Hsp90 via a central TPR domain, and features an intrinsically disordered C-terminal domain which is thought to bind hydrophobic substrates. Our focus lies in functional reconstitution and determining the structure of this complex. We are using X-ray crystallography, cryoEM, and an array of biophysical techniques, to unravel structural features that regulate the function of this vital complex.
Identification of chemical modulators of membrane protein proteostasis
We are screening small molecule libraries to identify novel inhibitors of membrane protein proteostasis. These molecules will serve as potent chemical probes to facilitate a comprehensive study of membrane protein proteostasis and the discovery of novel proteins, that regulate this process. In addition, our approach aims to identify lead compounds, paving the way for therapeutic development in areas where issues like membrane protein mislocalisation and protein misfolding contribute to disease. We employ high-throughput screening techniques utilising yeast and mammalian cells as model organisms.
Engineering membrane protein quality control systems for improved production of membrane proteins
The expression of eukaryotic membrane proteins in large quantities is often a major bottleneck to biochemical and structural studies. We are manipulating SGTA and the Bag6 complex to create mutants that minimise mislocalisation and degradation of membrane proteins during overexpression conditions. By investigating the targeting, integration, and function of these membrane proteins, our goal is to optimise production, yielding higher quantities of functional proteins. In addition, we are complementing this approach through mutagenesis of ribosomal proteins lining the ribosome exit tunnel to improve the expression of membrane proteins by targeting the ribosome itself.