Professor Ade Whitehouse
- Position
- Professor of Molecular Virology
- Areas of expertise
- Virology; RNA Biology, Epitransciptomics; Translational control, ncRNAs
- Phone
- +44(0)113 343 7096
- Location
- 6.44f Garstang
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
Introduction
Infection is a major cause of cancer worldwide. Viruses are associated with ~15% of human cancers, which approximates to about 2 million new cases every year in the world. Research in the Whitehouse laboratory aims to understand how viruses manipulate host cell pathways to replicate and cause cancer. We focus on studying the molecular biology of the two most recently discovered human tumour viruses, Kaposi's sarcoma-associated herpesvirus and Merkel cell polyomavirus. We utilise a range of cutting-edge transcriptomic and quantitative proteomic approaches to globally identify how viral proteins affect the host cell environment. This helps to identify essential virus-host cell interactions which we can then target by novel antiviral strategies to inhibit virus replication and transformation.
Current major projects
- Role of RNA modifications in virus infection
- Virus-induced specialised ribosomes
- Virus manipulation of ncRNA regulatory networks
- Development of novel therapeutic strategies to inhibit virus replication
Detailed research programme
Virus manipulation of m6A methylation
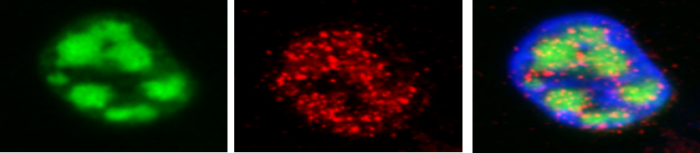
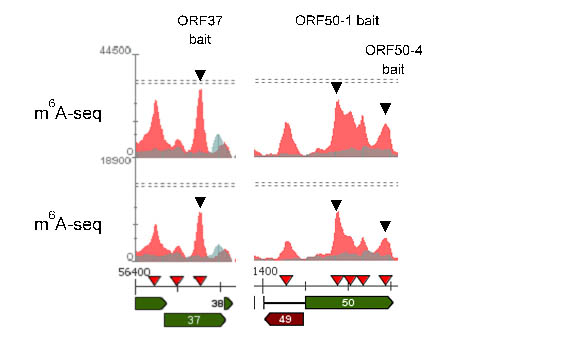
N6-methyladenosine (m6A) is the most abundant internal modification of cellular mRNAs. This dynamic modification is functionally linked to all stages of mRNA metabolism and regulates a variety of biological processes. As such, dysregulation of this pathway is implicated in the development of a wide range of diseases. We have recently show that m6A methylation has a pro-viral role enhancing KSHV replication. Current research aims to determine why viral transcripts get m6A methylated and the role of differentially methylated cellular transcripts during infection.
Virus-induced specialised ribosomes
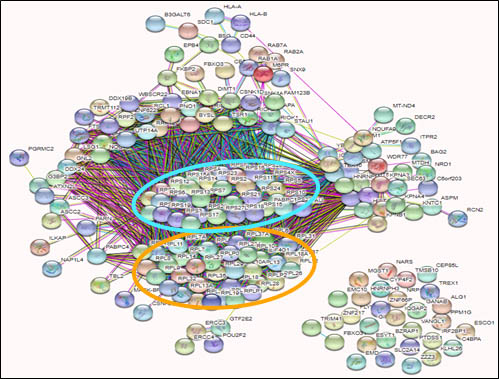
Ribosomes critical role in translating the genetic code, but were thought to have little influence on gene regulation. Evidence now reveals extensive variation between transcriptome expression and proteome levels, suggesting that cellular abundance of proteins may be controlled at a translational level. Aligned with this concept is emerging evidence of heterogeneity in ribosome composition leading to the formation of specialised ribosomes, which may have dedicated activities resulting in a substantial impact on how the genomic template is translated into functional proteins. We are examining how viruses may manipulate ribosome biogenesis to dictate how the host cell and viral genomic template is translated during infection.
Virus manipulation of host non-coding RNA regulatory networks
Non-coding RNAs (ncRNAs) constitute the majority of the human transcriptome. Emerging evidence suggests the existence of an unexpected interplay among ncRNA species. This intricate network of ncRNA:ncRNA interactions has the potential to modulate gene expression involved in multiple cellular pathways. As such, dysregulation of ncRNA regulatory networks may impact on the development and progression of a wide range of diseases. We are elucidating how ncRNA regulatory networks contribute to modulating gene expression, by determining how a virus manipulates the complex crosstalk between ncRNAs to regulate gene expression during infection. We are also exploring ncRNA regulatory networks as viable therapeutic targets to combat virus infection.
Development of novel therapeutic strategies to inhibit virus replication
Although vaccines have been developed for a few oncogenic viruses, these are not available for KSHV and MCPyV. Therefore novel antiviral strategies are required to combat these important human pathogens. Upon identification of essential virus-host cell interactions we utilise a structural-based rational drug design approach to molecular model and design small molecules to inhibit these interactions. Virtual high-throughput screening campaigns are conducted from a large libraries of commercially available compounds. Docking routines and ligand-similarity searches are utilised to design compounds which have the potential to inhibit these essential virus-host cell interactions. Once the virtual high-throughput screening campaign has been performed selected compounds are then assessed in virus-based assays for antiviral activity.