Professor David Rowlands
- Position
- Emeritus Professor of Molecular Virology
- Areas of expertise
- Virology, Structure, Cell Entry, Replication, Vaccines
- Location
- Garstang South 8.60
- Faculty
- Biological Sciences
- School
- Molecular and Cellular Biology
- Website
- ORCID
Introduction
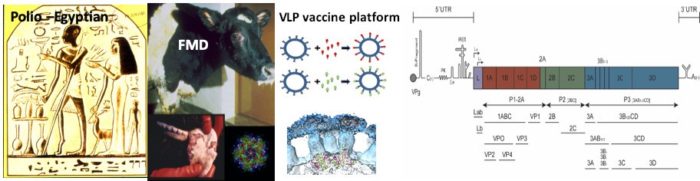
Picornaviruses are small single stranded positive sense RNA viruses and the aetiological agents of many important disease of man and livestock, including polio and foot-and-mouth disease (FMDV). Apart from their intrinsic interest as medical and veterinary pathogens they provide excellent subjects to study structure, biochemistry, cellular biology and immunology. Picornaviruses are amongst the smallest intracellular parasites and depend on close association with cell functions for all aspects of the infection process; cell entry, translation and replication, assembly and release. In addition to utilising cellular processes in their replication they also modulate the cellular environment to negate innate cellular immune mechanisms and maximise viral production.
Few options are available to control picornavirus infections. Drug development is difficult due to the small number of targetable functions consequent on their small genomes and their ability to mutate rapidly to avoid antiviral compounds. Vaccines have been more effective, but are available only for polio, FMD and hepatitis A. There are risks associated with conventional vaccines such as reversion to virulence by attenuated live vaccines and escape from containment facilities during manufacture of inactivated vaccines. There is great interest in the potential of recombinantly expressed virus-like particle (VLP) vaccines to obviate these risks.
Current major projects include
- Development of a novel ‘virus free’ polio VLP vaccine
- Development of a ‘universal’ VLP vaccine platform technology
- Mechanism(s) of cell entry by Picornaviruses
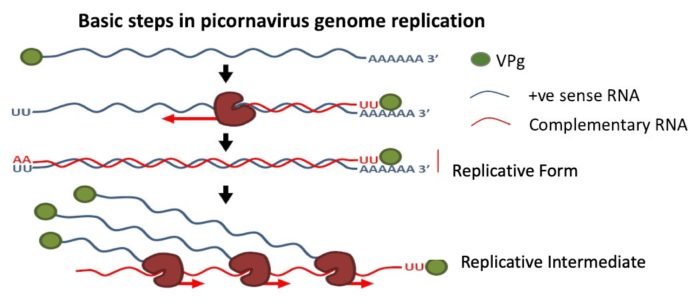
- Mechanisms of foot-and-mouth disease virus RNA replication
Detailed research programme
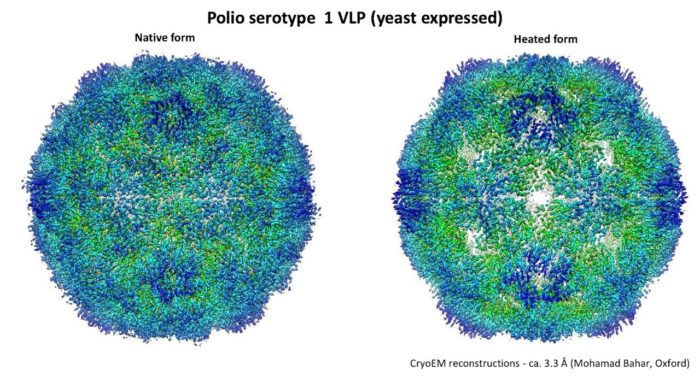
Stabilised poliovirus VLPs as safe replacements for inactivated polio vaccine (IPV).
With Professor Nic Stonehouse, I head a WHO funded international consortium to develop a safer polio vaccine.
Eradication of polio is an unachieved international goal. Live attenuated vaccines have played a major role in eliminating wild type virus but can revert to virulence. Inactivated polio vaccine is safer but requires growth of large quantities of virus during manufacture, with inherent risk of escape. We are developing recombinantly expressed VLPs as safer vaccines. These are antigenically indistinguishable from complete virus but are thermally unstable, converting to a conformation unsuitable as vaccine. This has been solved by introduction of mutations which produce stable VLPs. These are more amenable to industrial production and we anticipate partnering with industry to produce safer, ‘virus-free’ vaccines.
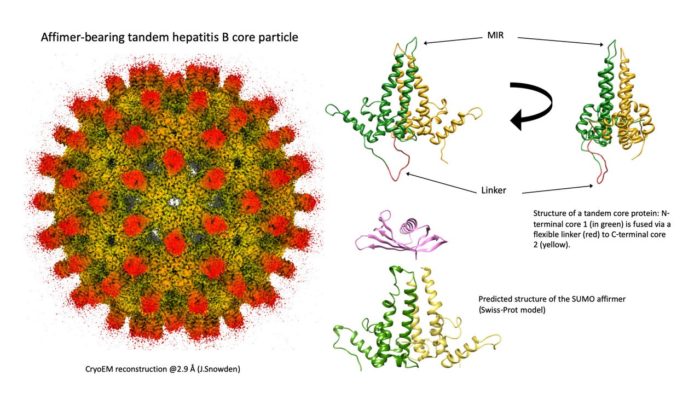
Modification of the self-assembling hepatitis B virus core protein to create novel VLP vaccine platform technology.
Many virus structural proteins self-assemble into highly immunogenic virus-like particles (VLPs) which can be modified as ‘platforms’ for presentation of protein antigens. Prof Stonehouse and I are investigating the core protein of hepatitis B virus (HBc) as a potential ‘universal’ VLP vaccine platform. HBc particles assemble from dimers of HBc protein and form prominent ‘spikes’ at the VLP surface. Affimers (synthetic antibody antigen binding domain mimics) inserted at the tips of these ‘spikes’ bind to their cognate sequences. Potential recombinant protein immunogens tagged with the affimer recognition sequence bind to the modified HBc VLPs to form immunogenic complexes. We are investigating the potential of this system for the development of novel vaccines, with special application for rapid responses to emerging infections.
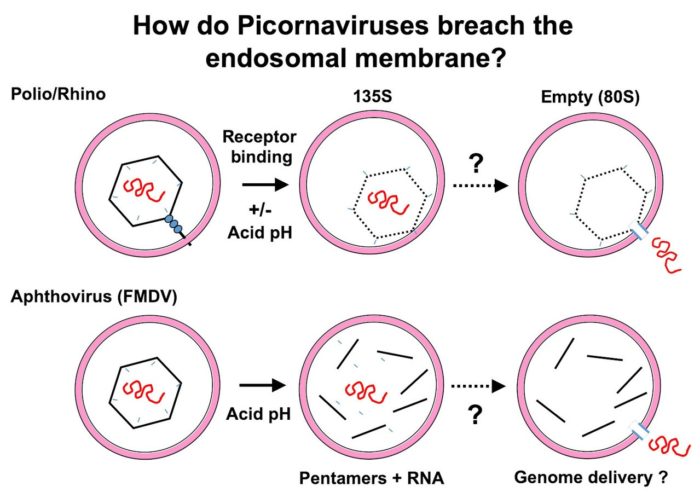
How do non-enveloped viruses such as picornaviruses introduce their RNA genomes into the cytoplasm?
The initial stages of infection by Picornaviruses require binding to receptors at the cell surface followed by internalisation into vesicles, such as endosomes. Subsequently, the RNA genome is transported across the endosomal membrane into the cytoplasm to initiate infection. Naked RNA is a labile molecule and highly susceptible to cleavage by nucleases, such as may be found in endosomal vesicles. We have shown that infection (and RNA integrity) is unaffected in the presence of high concentration of RNase despite co-internalisation into endosomes. How the RNA is protected during release for the virus particle and transfer across the endosomal membrane is poorly understood. We are investigating this problem in collaboration with colleagues in Oxford and Harvard.
What is the rationale for the unique features of the FMDV genome and their role in replication?
The foot-and-mouth disease virus (FMDV) genome resembles other members of the picornavirus family. However, a number of significant differences are unique to this virus. For example, the 5’ untranslated region (UTR) represents 1/7 of the genome and contains a number of highly structured and distinct RNA elements. No other picornaviruses have such large and complicated 5’UTRs and we are investigating the roles of these elements in FMDV replication. Another highly unusual feature is the presence of three copies of the peptide VPg, which primes RNA replication. Prof Stonehouse and I are investigating the roles of these elements in replication of viral RNA including the structure of the replication complex and the mechanisms controlling the switch from positive the negative sense RNA synthesis.